2. 上海图书馆(上海科学技术情报研究所), 上海 200031
2. Shanghai Library(Institute of Scientific and Technical Information of Shanghai), Shanghai 200031, China
2019年12月以来,湖北省武汉市陆续出现不明原因的肺炎病例,经各方研究,最终被确定为一种新型冠状病毒感染所致。为此,世界卫生组织将该病毒暂时命名为“2019新型冠状病毒(2019 novel coronavirus,2019-nCoV)”。2020年2月8日,国家卫生健康委员会决定将“新型冠状病毒感染的肺炎”暂命名为“新型冠状病毒肺炎”,简称“新冠肺炎(novel coronavirus pneumonia, NCP)”。NCP的肆虐严重威胁人类健康,给我国经济、社会带来了沉重的负担和严峻的挑战,与之相应的病毒溯源、传播机制和防控策略等方面的研究成为重大需求。NCP与历史上发生的严重急性呼吸综合征(severe acute respiratory syndrome, SARS)、中东呼吸综合征(Middle East respiratory syndrome, MERS)一样,均由冠状病毒(coronaviruses,CoVs)的感染引起。因而,针对CoVs已有研究进行梳理和总结,对当下NCP的防控研究或具有启示作用。
CoVs是外包有囊膜的单股正链RNA病毒,在系统分类上属巢状病毒目(Nidovirales)、冠状病毒科(Coronaviridae)、冠状病毒亚科(Coronavirinae)。2011年,国际病毒学分类委员会(International Committee on Taxonomy of Viruses, ICTV)将冠状病毒亚科又分为α、β、γ、δ 4个属。自1937年分离出第一种冠状病毒——禽类传染性支气管炎病毒(infectious bronchitis virus,IBV)以来,人类研究CoVs已有数十年的历史。2003年以来,严重急性呼吸综合征冠状病毒(severe acute respiratory syndrome coronaviruses,SARS-CoV)、中东呼吸综合征冠状病毒(Middle East respiratory syndrome coronaviruses,MERS-CoV)和2019-nCoV感染导致的疫情均引发了全世界的关注。
本文基于文献计量、引文分析和知识图谱等方法,就全球CoVs研究的学科和主题内容进行分析,以期总结全球CoVs研究的基本态势,为当下及今后CoVs感染所致疾病的疫情防控提供参考。
1 数据来源及分析 1.1 数据来源选取Web of Science®数据库作为主要的文献数据来源,以“冠状病毒”及其各个属为主题,在“Web of Science核心合集”中选择“高级检索”进行检索(检索时间为2020年1月28日),文献类型限定为“Article”“Review”和“Letter”,出版年限定为2000年至2019年。检索策略见表 1。
| 主题 | 检索式序号 | 检索式 | 文献量 |
| α冠状病毒 | #1 | (TS=(Alphacoronavirus OR "Alpaca coronavirus" OR "Alphacoronavirus 1" OR "Human coronavirus 229E" OR "HCoV-229E" OR "Human coronavirus NL63" OR "HCoV-NL63" OR "Miniopterus Bat coronavirus 1" OR "Bat CoV-1" OR "Bat-CoV-1A" OR "Bat-CoV-1B" OR "Miniopterus bat coronavirus HKU8" OR (Coronavirus AND HKU7) OR (Coronavirus AND HKU8) OR "Porcine epidemic diarrhea virus" OR "PED virus" OR (PEDV AND Coronavirus) OR "Rhinolophus Bat coronavirus HKU2" OR "RH-BAT-Cov-HKU2" OR "Scotophilus Bat coronavirus 512" OR " Sc-BatCoV-512" OR "Transmissible gastroenteritis virus" OR "Transmissible gastroenteritis coronavirus" OR (Coronavirus AND TGEV) OR "Feline coronavirus" OR "FCoV" OR "Feline infectious peritonitis" OR "Canine coronavirus" OR (Coronavirus AND CCoV)) AND PY=(2000-2019)) | 2 562 |
| β冠状病毒 | #2 | (TS=(Betacoronavirus OR "Betacoronavirus 1" OR "Human coronavirus HKU1" OR "HCoV-HKU1" OR "Murine coronavirus" OR "M-CoV" OR "rat coronavirus " OR (Coronavirus AND RtCoV) OR "Pipistrellus Bat coronavirus HKU5" OR "Bat-CoV HKU5" OR "Rousettus Bat coronavirus HKU9" OR "HKU9-1" OR "Severe acute respiratory syndrome" OR "SARS-CoV" OR " SARSr-CoV " OR (SARS and Coronavirus) OR "Tylonycteris Bat coronavirus HKU4" OR " Bat-CoV HKU4" OR " Middle East respiratory syndrome" OR "MERS-CoV" OR "2012-nCoV" OR "Human coronavirus OC43" OR "HCoV-OC43" OR "Hedgehog coronavirus" OR "EriCoV" OR "Wuhan coronavirus" OR "2019-nCoV" OR "2019 novel coronavirus" OR "Bovine coronavirus" OR (Coronavirus AND BCV) OR "equine coronavirus" OR "equine enteric coronavirus" OR (Coronavirus AND ECoV) OR "Porcine Hemagglutinating Encephalomyelitis" OR (Coronavirus AND PHEV) OR "canine respiratory coronavirus" OR (Coronavirus AND CrCoV) OR "Mouse Hepatitis Virus" OR (Coronavirus AND MHV) OR puffinosis OR "Puffinosis coronavirus" OR (Coronavirus AND PV)) AND PY=(2000-2019)) | 7 712 |
| γ冠状病毒 | #3 | (TS=(Gammacoronavirus OR "Avian coronavirus" OR "Beluga whale coronavirus" OR "Duck coronavirus" OR "Infectious bronchitis virus" OR (Coronavirus AND IBV) OR "Turkey coronavirus") AND PY=(2000-2019)) | 1 737 |
| δ冠状病毒 | #4 | (TS=(Deltacoronavirus OR "Bulbul coronavirus HKU11" OR (coronavirus AND HKU11) OR "Munia coronavirus HKU13" OR (coronavirus AND HKU13) OR "Thrush coronavirus HKU12" OR (coronavirus AND HKU12)) AND PY=(2000-2019)) | 133 |
| 冠状病毒 | #5 | ((TS=(coronavirus OR Coronavirinae) AND PY=(2000-2019)) OR #1 OR #2 OR #3 OR #4 | 13 025 |
采用文献计量、聚类分类等方法对文献数据进行分析,并运用引文分析和内容分析法进一步开展分析,阐述研究产出的分布、前沿和趋势。首先,运用文献计量法,对文献的年度分布、学科领域分布、主题分布等进行分析。其中,主题分析运用引文分析法,根据引证关系筛选出100篇高被引文献(其中2016年前的文献被引频次均为100次以上,2016—2019年的文献根据引证关系选择年度的高被引文献),并根据文献间的引证关系总结其研究主题。其次,通过内容分析与聚类分析,绘制CoVs研究的知识图谱。最后,基于国内外研究背景和热点的综合分析,梳理CoVs的主要研究动向,总结由其带来的启示。
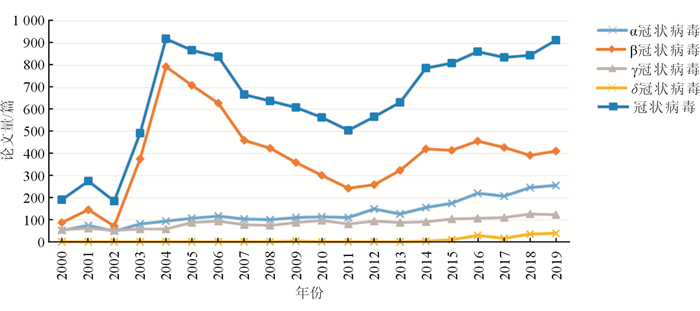
2 冠状病毒研究的论文产出 2.1 年度分布20世纪60年代,在人体内发现两种CoVs——人冠状病毒229E(human coronavirus 229E,HCoV-229E)
和人冠状病毒OC43(human coronavirus OC43,HCoV-OC43),分属于α冠状病毒和β冠状病毒,其感染所致疾病均为普通感冒。直到2003年前,在人体内发现的CoVs仍仅有这两种,因而全球CoVs的研究论文产出始终维持在较低的水平。2003年SARS暴发后,CoVs的研究在短期内成为焦点,随后逐年下降。2012年,MERS暴发后,CoVs再次引发关注,并在几年内再度成为全球病毒研究的热点。2003年之前,β冠状病毒的研究在α、β、γ、δ 4个属中最多,而SARS-CoV、MERS-CoV又均为β冠状病毒,因而全球CoVs的研究论文总量与β冠状病毒研究论文量变化趋势总体一致。2019年底发现的2019-nCoV也属β冠状病毒,可以预见,2020年全球β冠状病毒研究论文,以及CoVs的研究总体都将呈现快速增长的态势,论文发表量或将超2003年的规模(图 1)。
|
| 图 1 2000—2019年全球冠状病毒研究论文产出的年度变化 |
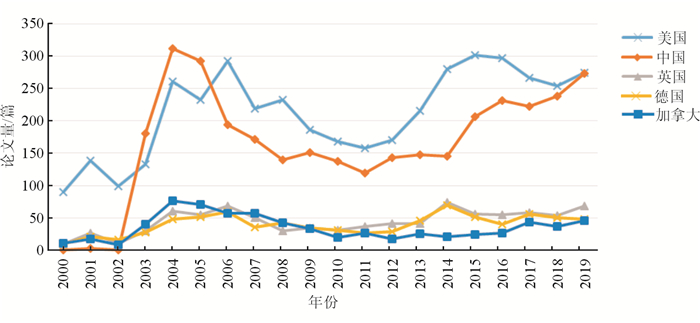
全球CoVs的研究以美国和中国较多。2003年SARS疫情暴发后,中国有关CoVs的研究论文快速增长,此后总体上与美国维持同一数量级的水平。近年来,美国的CoVs研究论文量基本在每年250~300篇,而中国的研究论文产出量整体呈上升的态势,至2019年,论文量已与美国大体相当(图 2)。其中,2019年中国的α冠状病毒研究的论文量(121篇)是美国(56篇)的2倍,而β冠状病毒的研究论文量则少于美国。2019年NCP暴发后,中国更加关注β冠状病毒的研究,预计研究论文量的增长趋势仍将延续。
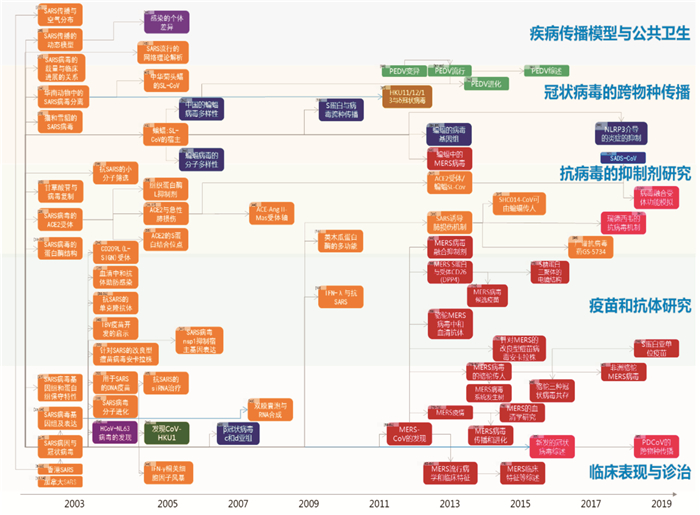
2.3 主题分布近20年来,一批高水平的研究论文得到发表,通过其引证关系可以了解CoVs研究主题的发展脉络(图 3)[1-100]。2003年对严重急性呼吸综合征的病原体SARS冠状病毒(SARS-CoV)的研究,是高被引论文发表的起点[1-4]。此后,研究者对SARS-CoV开展了基因组和分子进化方面的研究[15-17, 28],探讨SARS的动物传染源[23-24, 41-42],研究SARS传播模型及其公共卫生主题[21, 26-27, 43, 56],研发SARS疫苗[32-33, 40]和抗SARS的中和抗体[29],发现相关受体[25, 34-37]及小分子药物筛选[22, 38-39]。其中,SARS-CoV的传染源和贮存宿主的重点聚集于中华菊头蝠等蝙蝠[41-42]。继SARS-CoV之后,研究者分别于2004年和2005年发现了人冠状病毒NL63[54]和HKU1[55]。2012年12月发现了第6种人冠状病毒,2013年世界卫生组织将其命名为MERS-CoV[59-61]。自此,MERS-CoV研究相关的高被引论文[62-78]产出不断增加。除SARS-CoV和MERS-CoV外,多种其他CoVs研究也在近年来得到重视,如猪流行性腹泻病毒(PEDV)的变异和进化[86-91]即为热点之一。
|
| 图 3 2000—2019年全球冠状病毒研究高被引论文引证关系图 |
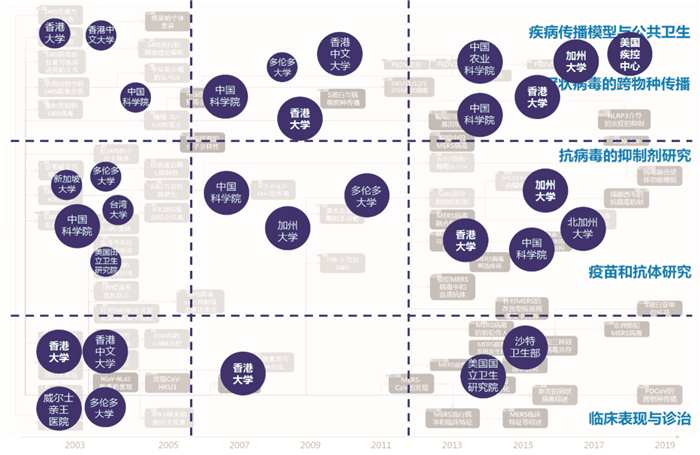
对SARS研究高峰期(2003—2005年)、SARS研究和MERS研究间隔期(2006—2011年)以及MERS研究和CoVs综合研究高峰期(2012—2019年)3个阶段的各个主题加以细分,分别统计各阶段、各主题的研究论文发表较多的机构,发现我国香港大学的研究较为系统,在各个阶段和各类主题均有涉及,是全球该领域的领先研究机构,另外还有中国科学院、加拿大多伦多大学、美国加州大学等。在人冠状病毒的临床研究方面,中国香港和沙特的研究机构,分别在SARS和MERS的研究中发表论文最多(图 4)。
|
| 图 4 2000—2019年全球冠状病毒研究论文的高产出机构分布 |
在已知的人类冠状病毒(HCoV)中,HCoV-NL63和HCoV-229E属于α冠状病毒属,HCoV-OC43、HCoV-HKU1、SARS-CoV、MERS-CoV以及2019-nCoV都属于β冠状病毒属[101]。在β冠状病毒属中,2019-nCoV被发现前,关注最多、开展相关研究最多的是SARS-CoV和MERS-CoV。以SARS-CoV、MERS-CoV相关研究的高被引论文内容分析为基础,绘制了冠状病毒研究的知识图谱(图 5)。

|
| 图 5 冠状病毒研究的知识图谱 |
病毒溯源和传播机制的研究是SARS-CoV和MERS-CoV早期研究的重点。研究表明,SARS-CoV和MERS-CoV的天然宿主可能都是蝙蝠[102],但是可能的中间宿主却很可能不同,因而SARS-CoV和MERS-CoV如何由动物向人类传播则是诸多研究的重点。从已有研究来看,果子狸和麝香猫可能是SARS-CoV的中间宿主,而MERS-CoV可能由单峰骆驼直接传播给人[103]。然而,从天然宿主到中间宿主,再到人传人传播,仍然有许多环节尚待进一步的流行病学和多学科研究加以综合阐明。反映在知识图谱上,关于冠状病毒的分子进化、传播机制和防控策略研究,需要做进一步的有机整合和集成。
目前,诸多的研究机构和企业也开展了抗病毒的小分子药物筛选以及单克隆抗体、疫苗等研究,其中靶向CoVs的表面刺突糖蛋白的研究较多,然而至今尚未成功开发出针对SARS-CoV和MERS-CoV的特效药物[104],且疫苗开发也面临不少的困难[105]。表现在知识图谱中,冠状病毒的多个蛋白与RNA的复合体结构研究仍有空白,而MERS-CoV感染的实验动物模型较少。虽然已建立了多个模拟SARS-CoV感染的动物模型,而且所构建的模型充分考虑了年龄等群体差异因素(如分别建立了年轻小鼠和衰老小鼠的模型),但MERS-CoV感染的啮齿动物(小鼠、仓鼠和豚鼠)模型和雪貂模型建立相对较难,这在很大程度上限制了关于MERS-CoV感染的抗病毒药物和疫苗的研发。
在SARS-CoV和MERS-CoV感染的发病机制研究方面,研究揭示CoVs蛋白的论文较多[106]。SARS-CoV和MERS-CoV的单正链RNA基因组都编码表面刺突糖蛋白(S)、包膜蛋白(E)、外膜蛋白(M)和核衣壳蛋白(N)。S蛋白是病毒表面三聚体糖蛋白,引导CoVs进入宿主细胞。SARS-CoV和MERS-CoV分别以人血管紧张素转换酶2(ACE2)和人二肽基肽酶(DPP4,又名CD26)作为主要受体。其中,ACE2广泛表达于肺泡、气管、支气管、支气管浆液腺、肺泡单核细胞和巨噬细胞,同时在动脉和静脉的内皮细胞、肠黏膜细胞、肾小管上皮细胞也有表达。免疫病理学的研究表明,SARS-CoV和MERS-CoV都可导致免疫紊乱[107]。SARS-CoV感染引发的细胞因子风暴[108]是导致患者病情加重乃至死亡的重要原因。因此,在尚无高效抗病毒药物的前提下,通过调节机体状态以提升免疫力,已是目前临床实践中的主要策略。在这方面,关于传统中医药在SARS防治中的实践应用有不少论文发表[109-110]。
4 展望与启示近20年来,全球冠状病毒研究力度明显提升,成果显著,对相关传染性疾病的防治和疫情防控发挥了重要支撑作用。虽然如此,人类对冠状病毒及其感染所致疾病的了解仍然有限,存在很多未知及空白,仍有待进一步提高,NCP的流行就是一个典型的例子。可喜的是,全球的科学家尤其是中国的科学家反应迅速,短时间内快速分离、鉴定出病原体并提出了有效的防控措施,后续关于2019-nCoV的病毒溯源、传播机制、病毒检测、疾病诊治乃至药物和疫苗研发以及疫情评估等方面的研究也正陆续开展。梳理既往包括SARS-CoV、MERS-CoV等的研究脉络,总结研究思路及路径,可对当前和未来冠状病毒的深入研究和传染病防控提供有益的启示。
4.1 从流行病学研究到疾病预警和防控策略的完善明确病毒的天然宿主和中间宿主,识别冠状病毒的系统进化特点,是科学有效地预警和防控策略制定的基础。当前,人们对SARS-CoV和MERS-CoV的天然宿主已有所了解,但对于SARS-CoV和MERS-CoV的传播途径仍尚未完全掌握,对于SARS-CoV、MERS-CoV和2019-nCoV由中间宿主向人传播的路径仍然有待进一步研究。在从人到人的传播方面,针对SARS-CoV和MERS-CoV的研究[27, 111]表明,室内环境的空气流动、动物内脏等废弃物的处理等细节,同样十分重要。此外,利用新技术、新方法完善病毒传播模型,对病毒传播的趋势预估在病毒防控中也十分必要。
4.2 从免疫病理学研究到冠状病毒感染患者的临床诊治由于缺乏特效的抗病毒药物,提高自身免疫力和对症治疗是应对当下新型冠状病毒肺炎患者的主要举措。目前,很多患者存在病情突然恶化或者好转后突然进展的现象,发病的免疫学机制尚不清楚,临床处置相当棘手,也无法预判,显著影响了患者预后。既往SARS-CoV、MERS-CoV感染患者诊治经验表明,很多重症患者体内会出现细胞因子风暴,给组织带来严重损伤,引发多器官功能衰竭。细胞因子风暴相关因子包括干扰素(IFN)、白细胞介素(IL)、趋化因子、集落刺激因子(CSF)、肿瘤坏死因子(TNF)等。目前,相关的研究已取得了不少进展,如研究发现IL-6可以作为判断疾病严重程度和预后指标的重要生物标志物[112]。然而,冠状病毒感染引发细胞因子风暴的信号转导途径尚未完全阐明,临床治疗方法和手段也有待进一步研究。
4.3 从结构生物学研究到抗病毒抑制剂的研发冠状病毒对宿主细胞的附着和进入、基因组转录、复制酶和结构蛋白的翻译等主要过程目前已经基本明确,这为抗病毒抑制剂的开发提供了依据。然而,从病毒的复制机制到抗病毒抑制剂的开发仍有许多难题有待攻克。首先,对于N蛋白-RNA、M蛋白-RNA复合物的结构了解不足[113],冠状病毒组装过程的研究仍有空白,这在一定程度上限制了抗病毒抑制剂的研究开发。其次,抗冠状病毒药物的研究开发中,广谱抗病毒药物的筛选和研究相对较多(如利巴韦林和瑞德西韦作为RNA聚合酶抑制剂已受到较多的关注),未来针对冠状病毒的精准靶向药物研发需要加强。再次,尽管针对SARS-CoV感染的实验动物模型构建已取得了诸多进展,但针对MARS-CoV感染的实验动物模型仍然有限。这也说明抗冠状病毒抑制剂开发所需的临床前研究仍存在一些短板,需要在未来的研究中加以关注。
4.4 从病毒免疫学研究到人冠状病毒疫苗的制备疫苗制备需要兼顾安全性、有效性和保护率,针对SARS-CoV的疫苗研发表明,结构蛋白编码基因中的变异给疫苗开发带来了难题。在冠状病毒的结构蛋白中,S蛋白是主要的抗原成分,重组载体疫苗、DNA疫苗和亚单位疫苗的很多开发以此为基础展开,其中又以基于受体结合区域(RBD)的疫苗研究最受关注。基于全长S蛋白的冠状病毒疫苗,可能会诱导有害免疫反应[114];基于S蛋白亚单位的疫苗,免疫原性又相对较低[103]。灭活病毒疫苗、减毒活病毒疫苗的开发也存在瓶颈。目前,针对SARS-CoV的疫苗评价已有研究,分别建立了基于年轻小鼠和衰老小鼠的模型。这提示人冠状病毒疫苗的开发需要充分考虑个体差异。因此,有待病毒免疫学的基础研究进一步取得突破,从而为人冠状病毒疫苗的开发提供更有力的支撑。已有较多针对猪、猫、狗、牛、家禽及其他鸟类的冠状病毒感染预防疫苗的研发,对其研究经验和效果评价的总结也可为针对人冠状病毒疫苗的开发提供一定的参考。
综上所述,重大突发传染病的防控是复杂的系统工程,需要建立跨学科、跨部门的传染病防控体系,才能使科学研究及其应用更为有序、有效。这有赖于基础医学、临床医学、公共卫生等相关学科的整合,有赖于科学研究与临床实践、疾病防控的协同,有赖于建立覆盖疾病防控、医疗卫生等多部门的全球数据信息共享和整合体系,从而为冠状病毒的防控提供更有力的支撑。面对冠状病毒的威胁,人类是命运共同体。相信在全球科学家的共同努力下,我们一定能有效防控新冠肺炎,一定能战胜冠状病毒感染引发的疫情。
编者按
2019年12月,首发于武汉的新型冠状病毒肺炎(novel coronavirus pneumonia, NCP)肆虐全国,严重威胁人类健康,引发了全球各界的密切关注。虽然近期已有一系列关于NCP的最新研究成果公布于世,但目前对于冠状病毒的认识仍存在诸多薄弱点、空白点,甚至是盲点。有关病毒的起源和宿主,传播感染途径,发病机制及变异机制,临床诊治的方向、思路和对策,疫苗的研制开发,以及防控措施的完善、高效应急体系的建立,均亟待开展跨学科、多机构、全方位的联合研究。这是一个事关全球人类命运共同体安全的重大科研命题,有赖于进一步加强全球合作,以取得突破性的研究进展。相信在全球科学家的共同努力下,在中国政府的有效干预下,我们一定能早日战胜新型冠状病毒肺炎疫情,造福全人类。
| [1] |
KSIAZEK T G, ERDMAN D, GOLDSMITH C S, et al. A novel coronavirus associated with severe acute respiratory syndrome[J]. N Engl J Med, 2003, 348(20): 1953-1966.
[DOI]
|
| [2] |
DROSTEN C, GVNTHER S, PREISER W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome[J]. N Engl J Med, 2003, 348(20): 1967-1976.
[DOI]
|
| [3] |
ROTA P A, OBERSTE M S, MONROE S S, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome[J]. Science, 2003, 300(5624): 1394-1399.
[DOI]
|
| [4] |
PEIRIS J S M, LAI S T, POON L L, et al. Coronavirus as a possible cause of severe acute respiratory syndrome[J]. Lancet, 2003, 361(9366): 1319-1325.
[DOI]
|
| [5] |
KUIKEN T, FOUCHIER R A, SCHUTTEN M, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome[J]. Lancet, 2003, 362(9380): 263-270.
[DOI]
|
| [6] |
RUAN Y J, WEI C L, EE A L, et al. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection[J]. Lancet, 2003, 361(9371): 1779-1785.
[DOI]
|
| [7] |
ZHONG N S, ZHENG B J, LI Y M, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003[J]. Lancet, 2003, 362(9393): 1353-1358.
[DOI]
|
| [8] |
LEE N, HUI D, WU A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong[J]. N Engl J Med, 2003, 348(20): 1986-1994.
[DOI]
|
| [9] |
TSANG K W, HO P L, OOI G C, et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong[J]. N Engl J Med, 2003, 348(20): 1977-1985.
[DOI]
|
| [10] |
DONNELLY C A, GHANI A C, LEUNG G M, et al. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong[J]. Lancet, 2003, 361(9371): 1761-1766.
[DOI]
|
| [11] |
NICHOLLS J M, POON L L, LEE K C, et al. Lung pathology of fatal severe acute respiratory syndrome[J]. Lancet, 2003, 361(9371): 1773-1778.
[DOI]
|
| [12] |
SETO W H, TSANG D, YUNG R W, et al. Effectiveness of precautions against droplets and contact in prevention of nosocomial transmission of severe acute respiratory syndrome (SARS)[J]. Lancet, 2003, 361(9368): 1519-1520.
[DOI]
|
| [13] |
POUTANEN S M, LOW D E, HENRY B, et al. Identification of severe acute respiratory syndrome in Canada[J]. N Engl J Med, 2003, 348(20): 1995-2005.
[DOI]
|
| [14] |
BOOTH C M, MATUKAS L M, TOMLINSON G A, et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area[J]. JAMA, 2003, 289(21): 2801-2809.
[DOI]
|
| [15] |
MARRA M A, JONES S J, ASTELL C R, et al. The genome sequence of the SARS-associated coronavirus[J]. Science, 2003, 300(5624): 1399-1404.
[DOI]
|
| [16] |
THIEL V, IVANOV K A, PUTICS A, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression[J]. J Gen Virol, 2003, 84(9): 2305-2315.
[DOI]
|
| [17] |
SNIJDER E J, BREDENBEEK P J, DOBBE J C, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage[J]. J Mol Biol, 2003, 331(5): 991-1004.
[DOI]
|
| [18] |
ANAND K, ZIEBUHR J, WADHWANI P, et al. Coronavirus main proteinase (3CLpro) structure:basis for design of anti-SARS drugs[J]. Science, 2003, 300(5626): 1763-1767.
[DOI]
|
| [19] |
YANG H, YANG M, DING Y, et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor[J]. Proc Natl Acad of Sci U S A, 2003, 100(23): 13190-13195.
[DOI]
|
| [20] |
CHOU K C, WEI D Q, ZHONG W Z. Binding mechanism of coronavirus main proteinase with ligands and its implication to drug design against SARS[J]. Biochem Biophys Res Commun, 2003, 308(1): 148-151.
[DOI]
|
| [21] |
PEIRIS J S, CHU C M, CHENG V C, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia:a prospective study[J]. Lancet, 2003, 361(9371): 1767-1772.
[DOI]
|
| [22] |
CINATL J, MORGENSTERN B, BAUER G, et al. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus[J]. Lancet, 2003, 361(9374): 2045-2046.
[DOI]
|
| [23] |
MARTINA B E, HAAGMANS B L, KUIKEN T, et al. Virology:SARS virus infection of cats and ferrets[J]. Nature, 2003, 425(6961): 915.
[DOI]
|
| [24] |
GUAN Y, ZHENG B J, HE Y Q, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China[J]. Science, 2003, 302(5643): 276-278.
[URI]
|
| [25] |
LI W, MOORE M J, VASILIEVA N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus[J]. Nature, 2003, 426(6965): 450-454.
[DOI]
|
| [26] |
LIPSITCH M, COHEN T, COOPER B, et al. Transmission dynamics and control of severe acute respiratory syndrome[J]. Science, 2003, 300(5627): 1966-1970.
[DOI]
|
| [27] |
LI Y, HUANG X, YU I T, et al. Role of air distribution in SARS transmission during the largest nosocomial outbreak in Hong Kong[J]. Indoor Air, 2005, 15(2): 83-95.
[DOI]
|
| [28] |
Chinese SARS Molecular Epidemiology Consortium. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China[J]. Science, 2004, 303(5664): 1666-1669.
[DOI]
|
| [29] |
SUBBARAO K, MCAULIFFE J, VOGEL L, et al. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice[J]. J Virol, 2004, 78(7): 3572-3577.
[DOI]
|
| [30] |
TRAGGIAI E, BECKER S, SUBBARAO K, et al. An efficient method to make human monoclonal antibodies from memory B cells:potent neutralization of SARS coronavirus[J]. Nat Med, 2004, 10(8): 871-875.
[DOI]
|
| [31] |
SUI J, LI W, MURAKAMI A, et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association[J]. Proc Natl Acad Sci U S A, 2004, 101(8): 2536-2541.
[DOI]
|
| [32] |
YANG Z Y, KONG W P, HUANG Y, et al. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice[J]. Nature, 2004, 428(6982): 561-564.
[DOI]
|
| [33] |
BISHT H, ROBERTS A, VOGEL L, et al. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice[J]. Proc Natl Acad Sci U S A, 2004, 101(17): 6641-6646.
[DOI]
|
| [34] |
JEFFERS S A, TUSELL S M, GILLIM-ROSS L, et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus[J]. Proc Natl Acad Sci U S A, 2004, 101(44): 15748-15753.
[DOI]
|
| [35] |
LI W, ZHANG C, SUI J, et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2[J]. EMBO J, 2005, 24(8): 1634-1643.
[DOI]
|
| [36] |
IMAI Y, KUBA K, RAO S, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure[J]. Nature, 2005, 436(7047): 112-116.
[DOI]
|
| [37] |
KUBA K, IMAI Y, RAO S, et al. A crucial role of angiotensin converting enzyme 2(ACE2) in SARS coronavirus-induced lung injury[J]. Nat Med, 2005, 11(8): 875-879.
[DOI]
|
| [38] |
SIMMONS G, GOSALIA D N, RENNEKAMP A J, et al. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry[J]. Proc Natl Acad Sci U S A, 2005, 102(33): 11876-11881.
[URI]
|
| [39] |
WU C Y, JAN J T, MA S H, et al. Small molecules targeting severe acute respiratory syndrome human coronavirus[J]. Proc Natl Acad Sci U S A, 2004, 101(27): 10012-10017.
[DOI]
|
| [40] |
CAVANAGH D. Severe acute respiratory syndrome vaccine development:experiences of vaccination against avian infectious bronchitis coronavirus[J]. Avian Pathol, 2003, 32(6): 567-582.
[DOI]
|
| [41] |
LI W, SHI Z, YU M, et al. Bats are natural reservoirs of SARS-like coronaviruses[J]. Science, 2005, 310(5748): 676-679.
[DOI]
|
| [42] |
LAU S K, WOO P C, LI K S, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats[J]. Proc Natl Acad Sci U S A, 2005, 102(39): 14040-14045.
[DOI]
|
| [43] |
MEYERS L A, POURBOHLOUL B, NEWMAN M E, et al. Network theory and SARS:predicting outbreak diversity[J]. J Theor Biol, 2005, 232(1): 71-81.
[DOI]
|
| [44] |
SANTOS R A, FERREIRA A J, SIMÕES E SILVA A C. Recent advances in the angiotensin-converting enzyme 2-angiotensin (1-7)-Mas axis[J]. Exp Physiol, 2008, 93(5): 519-527.
[DOI]
|
| [45] |
LI B J, TANG Q, CHENG D, et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque[J]. Nat Med, 2005, 11(9): 944-951.
[DOI]
|
| [46] |
HUANG K J, SU I J, THERON M, et al. An interferon-γ-related cytokine storm in SARS patients[J]. J Med Virol, 2005, 75(2): 185-194.
[DOI]
|
| [47] |
KAMITANI W, NARAYANAN K, HUANG C, et al. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation[J]. Proc Natl Acad Sci U S A, 2006, 103(34): 12885-12890.
[DOI]
|
| [48] |
WOO P C, WANG M, LAU S K, et al. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features[J]. J Virol, 2007, 81(4): 1574-1585.
[DOI]
|
| [49] |
KNOOPS K, KIKKERT M, WORM S H, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum[J]. PLoS Biol, 2008, 6(9): e226.
[DOI]
|
| [50] |
MORDSTEIN M, NEUGEBAUER E, DITT V, et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections[J]. J Virol, 2010, 84(11): 5670-5677.
[DOI]
|
| [51] |
CLEMENTZ M A, CHEN Z, BANACH B S, et al. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases[J]. J Virol, 2010, 84(9): 4619-4629.
[DOI]
|
| [52] |
TANG X C, ZHANG J X, ZHANG S Y, et al. Prevalence and genetic diversity of coronaviruses in bats from China[J]. J Virol, 2006, 80(15): 7481-7490.
[DOI]
|
| [53] |
WOO P C, LAU S K, LI K S, et al. Molecular diversity of coronaviruses in bats[J]. Virology, 2006, 351(1): 180-187.
[DOI]
|
| [54] |
WOO P C, LAU S K, CHU C M, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia[J]. J Virol, 2005, 79(2): 884-895.
[DOI]
|
| [55] |
VAN DER HOEK L, PYRC K, JEBBINK M F, et al. Identification of a new human coronavirus[J]. Nat Med, 2004, 10(4): 368-373.
[DOI]
|
| [56] |
LLOYD-SMITH J O, SCHREIBER S J, KOPP P E, et al. Superspreading and the effect of individual variation on disease emergence[J]. Nature, 2005, 438(7066): 355-359.
[DOI]
|
| [57] |
WOO P C, LAU S K, LAM C S, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus[J]. J Virol, 2012, 86(7): 3995-4008.
[DOI]
|
| [58] |
GRAHAM R L, BARIC R S. Recombination, reservoirs, and the modular spike:mechanisms of coronavirus cross-species transmission[J]. J Virol, 2010, 84(7): 3134-3146.
[DOI]
|
| [59] |
ZAKI A M, VAN BOHEEMEN S, BESTEBROER T M, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia[J]. N Engl J Med, 2012, 367(19): 1814-1820.
[DOI]
|
| [60] |
BERMINGHAM A, CHAND M A, BROWN C S, et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012[J]. Euro Surveill, 2012, 17(40): 20290.
|
| [61] |
MVLLER M A, RAJ V S, MUTH D, et al. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines[J]. mBio, 2012, 3(6): e00515-12.
|
| [62] |
ASSIRI A, AL-TAWFIQ J A, AL-RABEEAH A A, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia:a descriptive study[J]. Lancet Infect Dis, 2013, 13(9): 752-761.
[DOI]
|
| [63] |
MEMISH Z A, ZUMLA A I, AL-HAKEEM R F, et al. Family cluster of Middle East respiratory syndrome coronavirus infections[J]. N Engl J Med, 2013, 368(26): 2487-2494.
[DOI]
|
| [64] |
ZUMLA A, HUI D S, PERLMAN S. Middle East respiratory syndrome[J]. Lancet, 2015, 386(9997): 995-1007.
[DOI]
|
| [65] |
DE WIT E, VAN DOREMALEN N, FALZARANO D, et al. SARS and MERS:recent insights into emerging coronaviruses[J]. Nat Rev Microbiol, 2016, 14(8): 523-534.
[DOI]
|
| [66] |
LU L, LIU Q, ZHU Y, et al. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor[J]. Nat Commun, 2014, 5: 3067.
[DOI]
|
| [67] |
GE X Y, LI J L, YANG X L, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor[J]. Nature, 2013, 503(7477): 535-538.
[DOI]
|
| [68] |
LU G, HU Y, WANG Q, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26[J]. Nature, 2013, 500(7461): 227-231.
[DOI]
|
| [69] |
REUSKEN C B, HAAGMANS B L, MVLLER M A, et al. Middle East respiratory syndrome coronavirus neutra-lising serum antibodies in dromedary camels:a comparative serological study[J]. Lancet Infect Dis, 2013, 13(10): 859-866.
[DOI]
|
| [70] |
ASSIRI A, MCGEER A, PERL T M, et al. Hospital outbreak of Middle East respiratory syndrome coronavirus[J]. N Engl J Med, 2013, 369(5): 407-416.
[DOI]
|
| [71] |
DROSTEN C, MEYER B, MVLLER M A, et al. Transmission of MERS-coronavirus in household contacts[J]. N Engl J Med, 2014, 371(9): 828-835.
[DOI]
|
| [72] |
AL-ABDALLAT M M, PAYNE D C, ALQASRAWI S, et al. Hospital-associated outbreak of Middle East respiratory syndrome coronavirus:a serologic, epidemiologic, and clinical description[J]. Clin Infect Dis, 2014, 59(9): 1225-1233.
[DOI]
|
| [73] |
WALLS A C, TORTORICI M A, BOSCH B J, et al. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer[J]. Nature, 2016, 531(7592): 114-117.
[DOI]
|
| [74] |
HAAGMANS B L, VAN DEN BRAND J M A, RAJ V S, et al. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels[J]. Science, 2016, 351(6268): 77-81.
[DOI]
|
| [75] |
WANG L, SHI W, JOYCE M G, et al. Evaluation of candidate vaccine approaches for MERS-CoV[J]. Nat Commun, 2015, 6: 7712.
[DOI]
|
| [76] |
MVLLER M A, MEYER B, CORMAN V M, et al. Presence of Middle East respiratory syndrome coronavirus antibodies in Saudi Arabia:a nationwide, cross-sectional, serological study[J]. Lancet Infect Dis, 2015, 15(6): 629.
[DOI]
|
| [77] |
COTTEN M, WATSON S J, ZUMLA A I, et al. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus[J]. mBio, 2014, 5(1): e01062-13.
|
| [78] |
DROSTEN C, SEILMAIER M, CORMAN V M, et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection[J]. Lancet Infect Dis, 2013, 13(9): 745-751.
[DOI]
|
| [79] |
HAAGMANS B L, AL DHAHIRY S H, REUSKEN C B, et al. Middle East respiratory syndrome coronavirus in dromedary camels:an outbreak investigation[J]. Lancet Infect Dis, 2014, 14(2): 140-145.
[DOI]
|
| [80] |
AZHAR E I, EL-KAFRAWY S A, FARRAJ S A, et al. Evidence for camel-to-human transmission of MERS coronavirus[J]. N Engl J Med, 2014, 370(26): 2499-2505.
|
| [81] |
MEMISH Z A, COTTEN M, MEYER B, et al. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013[J]. Emerg Infect Dis, 2014, 20(6): 1012-1015.
[DOI]
|
| [82] |
CORMAN V M, ITHETE N L, RICHARDS L R, et al. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat[J]. J Virol, 2014, 88(19): 11297-11303.
[DOI]
|
| [83] |
SABIR J S, LAM T T, AHMED M M, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia[J]. Science, 2016, 351(6268): 81-84.
[DOI]
|
| [84] |
ZHANG G, COWLED C, SHI Z, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity[J]. Science, 2013, 339(6118): 456-460.
[URI]
|
| [85] |
ANNAN A, BALDWIN H J, CORMAN V M, et al. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe[J]. Emerg Infect Dis, 2013, 19(3): 456-459.
[DOI]
|
| [86] |
LI W, LI H, LIU Y, et al. New variants of porcine epidemic diarrhea virus, China, 2011[J]. Emerg Infect Dis, 2012, 18(8): 1350-1353.
[DOI]
|
| [87] |
STEVENSON G W, HOANG H, SCHWARTZ K J, et al. Emergence of porcine epidemic diarrhea virus in the United States:clinical signs, lesions, and viral genomic sequences[J]. J Vet Diagn Invest, 2013, 25(5): 649-654.
[DOI]
|
| [88] |
HUANG Y W, DICKERMAN A W, PIÑEYRO P, et al. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States[J]. mBio, 2013, 4(5): e00737-13.
[URI]
|
| [89] |
SUN R Q, CAI R J, CHEN Y Q, et al. Outbreak of porcine epidemic diarrhea in suckling piglets, China[J]. Emerg Infect Dis, 2012, 18(1): 161-163.
[DOI]
|
| [90] |
VLASOVA A N, MARTHALER D, WANG Q, et al. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014[J]. Emerg Infect Dis, 2014, 20(10): 1620-1628.
|
| [91] |
LEE C. Porcine epidemic diarrhea virus:an emerging and re-emerging epizootic swine virus[J]. Virol J, 2015, 12: 193.
[DOI]
|
| [92] |
ZHOU P, FAN H, LAN T, et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin[J]. Nature, 2018, 556(7700): 255-258.
[DOI]
|
| [93] |
GRALINSKI L E, BANKHEAD A 3RD, JENG S, et al. Mechanisms of severe acute respiratory syndrome coronavirus-induced acute lung injury[J]. mBio, 2013, 4(4): e00271-13.
[URI]
|
| [94] |
AGOSTINI M L, ANDRES E L, SIMS A C, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease[J]. mBio, 2018, 9(2): e00221-18.
|
| [95] |
LI W, HULSWIT R J G, KENNEY S P, et al. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility[J]. Proc Natl Acad Sci U S A, 2018, 115(22): E5135-E5143.
[DOI]
|
| [96] |
SHEAHAN T P, SIMS A C, GRAHAM R L, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses[J]. Sci Transl Med, 2017, 9(396): eaal3653.
[DOI]
|
| [97] |
MENACHERY V D, YOUNT B L JR, DEBBINK K, et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence[J]. Nat Med, 2015, 21(12): 1508-1513.
[DOI]
|
| [98] |
ADNEY D R, WANG L, VAN DOREMALEN N, et al. Efficacy of an adjuvanted Middle East respiratory syndrome coronavirus spike protein vaccine in dromedary camels and alpacas[J]. Viruses, 2019, 11(3): E212.
[DOI]
|
| [99] |
AHN M, ANDERSON D E, ZHANG Q, et al. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host[J]. Nat Microbiol, 2019, 4(5): 789-799.
[DOI]
|
| [100] |
WALLS A C, XIONG X, PARK Y J, et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion[J]. Cell, 2019, 176(5): 1026-1039. e15.
[DOI]
|
| [101] |
AL-TAWFIQ J A, AUWAERTER P G. Healthcare-associated infections:the hallmark of Middle East respiratory syndrome coronavirus with review of the literature[J]. J Hosp Infect, 2019, 101(1): 20-29.
[DOI]
|
| [102] |
CUI J, LI F, SHI Z L. Origin and evolution of pathogenic coronaviruses[J]. Nat Rev Microbiol, 2019, 17(3): 181-192.
[DOI]
|
| [103] |
SONG Z, XU Y, BAO L, et al. From SARS to MERS, thrusting coronaviruses into the spotlight[J]. Viruses, 2019, 11(1): 59.
[DOI]
|
| [104] |
SHEN L, NIU J, WANG C, et al. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses[J]. J Virol, 2019, 93(12): e00023-e00019.
|
| [105] |
MODJARRAD K, ROBERTS C C, MILLS K T, et al. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine:a phase 1, open-label, single-arm, dose-escalation trial[J]. Lancet Infect Dis, 2019, 19(9): 1013-1022.
[DOI]
|
| [106] |
XIA S, YAN L, XU W, et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike[J]. Sci Adv, 2019, 5(4): eaav4580.
[DOI]
|
| [107] |
SAIF L J, WANG Q, VLASOVA A N, et al. Diseases of Swine[M]. 11th. New York: Wiley Blackwell, 2019: 488-523.
|
| [108] |
CHANNAPPANAVAR R, PERLMAN S. Pathogenic human coronavirus infections:causes and consequences of cytokine storm and immunopathology[J]. Semin Immunopathol, 2017, 39(5): 529-539.
[DOI]
|
| [109] |
颜德馨, 余小萍, 石克华, 等. 传染性非典型性肺炎的中医证治探讨[J]. 中西医结合学报, 2004, 2(4): 241-244. [URI]
|
| [110] |
袁长津, 葛金文, 周慎, 等. 中医对SARS的认识及辨证治疗方案——92篇中医药防治SARS文献研究小结[J]. 中国医药学报, 2004, 19(6): 371-373. [URI]
|
| [111] |
HEMIDA M G, ALNAEEM A. Some One Health based control strategies for the Middle East respiratory syndrome coronavirus[J]. One Health, 2019, 8: 100102.
[DOI]
|
| [112] |
TANAKA T, NARAZAKI M, KISHIMOTO T. Immunotherapeutic implications of IL-6 blockade for cytokine storm[J]. Immunotherapy, 2016, 8(8): 959-970.
[DOI]
|
| [113] |
MASTERS P S. Coronavirus genomic RNA packaging[J]. Virology, 2019, 537: 198-207.
[DOI]
|
| [114] |
CZUB M, WEINGARTL H, CZUB S, et al. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets[J]. Vaccine, 2005, 23(17-18): 2273-2279.
[DOI]
|
 2020, Vol. 27
2020, Vol. 27